Preprocessing time series with aeon¶
It is common to need to preprocess time series data before applying machine learning algorithms. So algorithms can handle these characteristics, or aeon transformers can be used to preprocess collections of time series into standard format. This notebook demonstrates three common use cases
Rescaling time series¶
Different levels of scale and variance can mask discriminative patterns in time series. This is particularly true for methods that are based on distances. It common to rescale time series to have zero mean and unit variance. For example, the data in the UnitTest dataset is a subset of the [Chinatown dataset] (https://timeseriesclassification.com/description.php?Dataset=Chinatown. These are counts of pedestrians in Chinatown, Melbourne. The time series are of different means
[3]:
import numpy as np
from aeon.datasets import load_unit_test
X, y = load_unit_test(split="Train")
np.mean(X, axis=-1)[0:5]
[3]:
array([[561.875 ],
[604.95833333],
[629.16666667],
[801.45833333],
[540.75 ]])
[4]:
np.std(X, axis=-1)[0:5]
[4]:
array([[428.95224215],
[483.35481095],
[514.90052977],
[629.00847763],
[389.10059218]])
We can rescale the time series in three ways:
Normalise: subtract the mean and divide by the standard deviation to make all series have zero mean and unit variance.
[5]:
from aeon.transformations.collection import Normalizer
normalizer = Normalizer()
X2 = normalizer.fit_transform(X)
np.round(np.mean(X2, axis=-1)[0:5], 6)
[5]:
array([[ 0.],
[-0.],
[ 0.],
[-0.],
[-0.]])
[6]:
np.round(np.std(X2, axis=-1)[0:5], 6)
[6]:
array([[1.],
[1.],
[1.],
[1.],
[1.]])
Re-center: Recentering involves subtracting the mean of each series
[7]:
from aeon.transformations.collection import Centerer
c = Centerer()
X3 = c.fit_transform(X)
np.round(np.mean(X3, axis=-1)[0:5], 6)
[7]:
array([[ 0.],
[-0.],
[ 0.],
[-0.],
[ 0.]])
Min-Max: Scale the data to be between 0 and 1
[8]:
from aeon.transformations.collection import MinMaxScaler
minmax = MinMaxScaler()
X4 = minmax.fit_transform(X)
np.round(np.min(X4, axis=-1)[0:5], 6)
[8]:
array([[0.],
[0.],
[0.],
[0.],
[0.]])
[9]:
np.round(np.max(X4, axis=-1)[0:5], 6)
[9]:
array([[1.],
[1.],
[1.],
[1.],
[1.]])
There is no best way to do this, although for counts such as this it is more common to MinMax scale, so that the data still has some interpretation as proportions.
Resizing time series¶
Suppose we have a collections of time series with different lengths, i.e. different number of time points. Currently, most of aeon’s collection estimators (classification, clustering or regression) require equal-length time series. Those that can handle unequal length series are tagged with “capability:unequal”.
[10]:
from aeon.classification.convolution_based import RocketClassifier
from aeon.datasets import load_basic_motions, load_japanese_vowels, load_plaid
from aeon.utils.validation import has_missing, is_equal_length, is_univariate
If you want to use an estimator that cannot internally handle missing values, one option is to convert unequal length series into equal length. This can be done through padding, truncation or resizing through fitting a function and resampling.
Unequal or equal length collections time series¶
If a collection contains all equal length series, it will store the data in a 3D numpy of shape (n_cases, n_channels, n_timepoints). If it is unequal length, it is stored in a list of 2D numpy arrays:
[11]:
# Equal length multivariate data
bm_X, bm_y = load_basic_motions()
X = bm_X
print(f"{type(X)}, {X.shape}")
print(
f"univariate = {is_univariate(X)}, has missing ={has_missing(X)}, equal "
f"length = {is_equal_length(X)}"
)
<class 'numpy.ndarray'>, (80, 6, 100)
univariate = False, has missing =False, equal length = True
[12]:
# Unequal length univariate data
plaid_X, plaid_y = load_plaid()
X = plaid_X
print(type(plaid_X), "\n", plaid_X[0].shape, "\n", plaid_X[10].shape)
print(
f"univariate = {is_univariate(X)}, has missing ={has_missing(X)}, equal "
f"length = {is_equal_length(X)}"
)
<class 'list'>
(1, 500)
(1, 300)
univariate = True, has missing =False, equal length = False
[13]:
vowels_X, vowels_y = load_japanese_vowels(split="train")
X = vowels_X
print(
f"univariate = {is_univariate(X)}, has missing ={has_missing(X)}, equal "
f"length = {is_equal_length(X)}"
)
univariate = False, has missing =False, equal length = False
[14]:
series_lengths = [array.shape[1] for array in plaid_X]
# Find the minimum and maximum of the second dimensions
min_length = min(series_lengths)
max_length = max(series_lengths)
print(" Min length = ", min_length, " max length = ", max_length)
Min length = 100 max length = 1344
There are two basic strategies for unequal length problems
Use an estimator that can internally handle missing values
Transform the data to be equal length by, for example, truncating or padding series
Estimators with the tag "capability:unequal_length": True have the capability to handle unequal length series. For classification, regression and clusterign, the current list is
[15]:
from aeon.utils.discovery import all_estimators
all_estimators(
type_filter=["classifier", "regressor", "clusterer"],
tag_filter={"capability:unequal_length": True},
)
[15]:
[('Catch22Classifier',
aeon.classification.feature_based._catch22.Catch22Classifier),
('Catch22Clusterer', aeon.clustering.feature_based._catch22.Catch22Clusterer),
('Catch22Regressor', aeon.regression.feature_based._catch22.Catch22Regressor),
('DummyClassifier', aeon.classification.dummy.DummyClassifier),
('DummyRegressor', aeon.regression._dummy.DummyRegressor),
('ElasticEnsemble',
aeon.classification.distance_based._elastic_ensemble.ElasticEnsemble),
('KNeighborsTimeSeriesClassifier',
aeon.classification.distance_based._time_series_neighbors.KNeighborsTimeSeriesClassifier),
('KNeighborsTimeSeriesRegressor',
aeon.regression.distance_based._time_series_neighbors.KNeighborsTimeSeriesRegressor),
('RDSTClassifier', aeon.classification.shapelet_based._rdst.RDSTClassifier),
('RDSTRegressor', aeon.regression.shapelet_based._rdst.RDSTRegressor)]
You can pass these estimators unequal length series and they will work as expected.
[16]:
from aeon.classification.distance_based import KNeighborsTimeSeriesClassifier
knn = KNeighborsTimeSeriesClassifier()
model = knn.fit(plaid_X, plaid_y)
If time series are unequal length, collection estimators will raise an error if they do not have the capability to handle this characteristic. If you want to use them, you will need to preprocess the data to be equal length.
[17]:
rc = RocketClassifier()
try:
rc.fit(plaid_X, plaid_y)
except ValueError as e:
print(f"ValueError: {e}")
ValueError: Data seen by instance of RocketClassifier has unequal length series, but RocketClassifier cannot handle unequal length series.
Padding, truncating or resizing.¶
We can pad, truncate or resize. By default, pad adds zeros to make all series the length of the longest, truncate removes all values beyond the length of the shortest and resize stretches or shrinks the series.
[18]:
from aeon.transformations.collection import Padder, Resizer, Truncator
pad = Padder()
truncate = Truncator()
resize = Resizer(length=600)
X2 = pad.fit_transform(plaid_X)
X3 = truncate.fit_transform(plaid_X)
X4 = resize.fit_transform(plaid_X)
print(X2.shape, "\n", X3.shape, "\n", X4.shape)
(1074, 1, 1344)
(1074, 1, 100)
(1074, 1, 600)
[19]:
import matplotlib.pyplot as plt
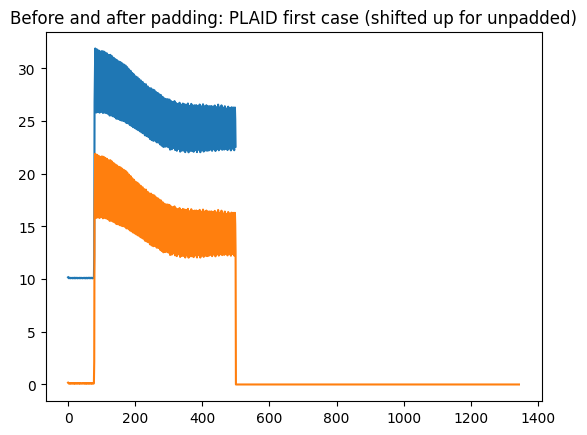
plt.title("Before and after padding: PLAID first case (shifted up for unpadded)")
plt.plot(plaid_X[0][0] + 10)
plt.plot(X2[0][0])
[19]:
[<matplotlib.lines.Line2D at 0x20069acd130>]
You can put these transformers in a pipeline to apply to both train/test split
[20]:
from sklearn.metrics import accuracy_score
# Unequal length univariate data
from aeon.pipeline import make_pipeline
train_X, train_y = load_plaid(split="Train")
test_X, test_y = load_plaid(split="Test")
steps = [truncate, rc]
pipe = make_pipeline(steps)
pipe.fit(train_X, train_y)
preds = pipe.predict(test_X)
accuracy_score(train_y, preds)
[20]:
0.813780260707635
Missing Values¶
Missing values are indicated by NaN in numpy array. You can test whether any aeon data structure contains missing values using the utility function
[21]:
X = np.random.random(size=(10, 2, 200))
has_missing(X)
[21]:
False
[22]:
X[5][0][55] = np.NAN
has_missing(X)
[22]:
True
There are a range of strategies for handling missing values. These include:
Use an estimator that internally handles missing values. It is fairly easy for some algorithms (such as decision trees) to internally deal with missing values, usually be using it as a distinct series value after discretisation. We do not yet have many estimators with this capability. Estimators that are able to internally handle missing values are tagged with
"capability:missing_values": True.
[23]:
from aeon.utils.discovery import all_estimators
all_estimators(
tag_filter={"capability:missing_values": True},
)
[23]:
[('BORF', aeon.transformations.collection.dictionary_based._borf.BORF),
('CollectionId',
aeon.transformations.collection.compose._identity.CollectionId),
('DummyClassifier', aeon.classification.dummy.DummyClassifier),
('DummyRegressor', aeon.regression._dummy.DummyRegressor),
('RandomSegmenter', aeon.segmentation._random.RandomSegmenter),
('STRAY', aeon.anomaly_detection._stray.STRAY),
('SimpleImputer', aeon.transformations.collection._impute.SimpleImputer)]
Removing series with missing: this is often desirable if the train set size is large, the number of series with missing is small and the proportion of missing values for these series is high.
We do not yet have a transformer for this, but it is easy to implement yourself.
Interpolating missing values from series: estimating the missing values from the other values in a time series is commonly done. This is often desirable if the train set size is small and the proportion of missing values is low. You can do this with the transformer
SimpleImputer. This interpolates each series and each channel independently. So for example a mean interpolation of series with two channels[[NaN,1.0,2.0,3.0],[-1.0,-2.0,-3.0,-4.0]]would be[[2.0,1.0,2.0,3.0],[-1.0,-2.0,-3.0,-4.0]].
[26]:
from aeon.transformations.collection import SimpleImputer
imput = SimpleImputer(strategy="mean")
X2 = imput.fit_transform(X)
has_missing(X2)
[26]:
False
[27]:
imp2 = SimpleImputer(strategy="median")
X3 = imp2.fit_transform(X)
has_missing(X3)
[27]:
False
[28]:
imp3 = SimpleImputer(strategy="constant", fill_value=0)
X4 = imp3.fit_transform(X)
has_missing(X4)
[28]:
False
[ ]:
Generated using nbsphinx. The Jupyter notebook can be found here.