Benchmarking: retrieving and comparing against published results¶
You can access all archived published results for time series classification (TSC) directly with aeon. These results are all stored on the website timeseriesclassification.com.
These are reference results tied to publications and will not change. The datasets and estimators for recovering these results are intentionally hard coded and not generalised, to remove any potential for confusion. To more flexibly load the latest results for classification, clustering and regression, see the notebook on loading reference results.
We compare results with the critical difference graph described in this notebook. Note that the way we group classifiers has slightly changed and hence there may be small variation in cliques from published results.
The published results can be recovered from the time series classifcation website directly or with aeon.
Classification¶
These results were presented in three bake offs for classification: The first bake off [1] used 85 UCR univariate TSC datasets. The second bake off [2] introduced the multivariate TSC archive, and compared classifier performance. The third bake off [3], the bake off redux, compared univariate classifiers on 112 UCR datasets.
The great time series classification bake off, 2017¶
The first TSC bake off [1], conducted in 2015 and published in 2017 compared 25 classifiers on the 85 UCR data that were released in 2015. The publication is available here.
You can pull down results for the original bake off using the following function. The default train/test split is returned as the first resample, and there are 100 resamples available for most experiments. The data resampling function used is not the same as the one available in aeon.
[1]:
from aeon.benchmarking.published_results import (
load_classification_bake_off_2017_results,
)
results_dict = load_classification_bake_off_2017_results(num_resamples=10)
results_dict["FlatCOTE"]["GunPoint"]
[1]:
array([1. , 0.98666667, 0.99333333, 1. , 1. ,
0.97333333, 0.98 , 0.99333333, 1. , 1. ])
We were unable to recover experiment resamples past a certain point for some classifier/dataset combinations. Missing resamples will return a NaN
[2]:
results_dict["DTW_F"]["FordB"]
[2]:
array([0.74938272, 0.88888889, 0.78765432, 0.88148148, 0.87407407,
0.87654321, nan, nan, nan, nan])
[3]:
results_arr, datasets, classifiers = load_classification_bake_off_2017_results(
num_resamples=100, as_array=True, ignore_nan=True
)
results_arr.shape
[3]:
(85, 25)
The dataset used for the first bake off are described in [4]:
[4]:
datasets
[4]:
['Adiac',
'ArrowHead',
'Beef',
'BeetleFly',
'BirdChicken',
'Car',
'CBF',
'ChlorineConcentration',
'CinCECGTorso',
'Coffee',
'Computers',
'CricketX',
'CricketY',
'CricketZ',
'DiatomSizeReduction',
'DistalPhalanxOutlineCorrect',
'DistalPhalanxOutlineAgeGroup',
'DistalPhalanxTW',
'Earthquakes',
'ECG200',
'ECG5000',
'ECGFiveDays',
'ElectricDevices',
'FaceAll',
'FaceFour',
'FacesUCR',
'FiftyWords',
'Fish',
'FordA',
'FordB',
'GunPoint',
'Ham',
'HandOutlines',
'Haptics',
'Herring',
'InlineSkate',
'InsectWingbeatSound',
'ItalyPowerDemand',
'LargeKitchenAppliances',
'Lightning2',
'Lightning7',
'Mallat',
'Meat',
'MedicalImages',
'MiddlePhalanxOutlineCorrect',
'MiddlePhalanxOutlineAgeGroup',
'MiddlePhalanxTW',
'MoteStrain',
'NonInvasiveFetalECGThorax1',
'NonInvasiveFetalECGThorax2',
'OliveOil',
'OSULeaf',
'PhalangesOutlinesCorrect',
'Phoneme',
'Plane',
'ProximalPhalanxOutlineCorrect',
'ProximalPhalanxOutlineAgeGroup',
'ProximalPhalanxTW',
'RefrigerationDevices',
'ScreenType',
'ShapeletSim',
'ShapesAll',
'SmallKitchenAppliances',
'SonyAIBORobotSurface1',
'SonyAIBORobotSurface2',
'StarLightCurves',
'Strawberry',
'SwedishLeaf',
'Symbols',
'SyntheticControl',
'ToeSegmentation1',
'ToeSegmentation2',
'Trace',
'TwoLeadECG',
'TwoPatterns',
'UWaveGestureLibraryX',
'UWaveGestureLibraryY',
'UWaveGestureLibraryZ',
'UWaveGestureLibraryAll',
'Wafer',
'Wine',
'WordSynonyms',
'Worms',
'WormsTwoClass',
'Yoga']
The classifiers used are as follows:
[5]:
classifiers
[5]:
['ACF',
'BOSS',
'CID_DTW',
'CID_ED',
'DDTW_R1_1NN',
'DDTW_Rn_1NN',
'DTW_F',
'EE',
'ERP_1NN',
'Euclidean_1NN',
'FlatCOTE',
'FS',
'LCSS_1NN',
'LPS',
'LS',
'MSM_1NN',
'PS',
'RotF',
'SAXVSM',
'ST',
'TSBF',
'TSF',
'TWE_1NN',
'WDDTW_1NN',
'WDTW_1NN']
Once you have the results you want, you can compare classifiers with built in aeon tools.
Suppose we want to recreate the critical difference diagram published in [1]:
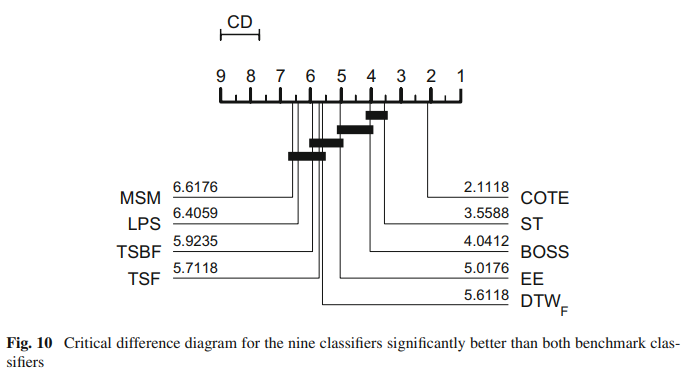
This displays the critical difference diagram [6] for comparing classifiers. It shows the average rank of each estimator over all datasets. It then groups estimators for which there is no significant difference in rank into cliques, shown with a solid bar. The published results used the original method for finding cliques called the post hoc Nemenyi test. Our plotting tool offers this as an alternative. See the docs for aeon.visualisation.plot_critical_difference for more
details. To recreate the above, we can do this (note slight difference in names, MSM_1NN is MSM and FlatCOTE is COTE.
[6]:
from aeon.visualisation import plot_critical_difference
subsample = ["MSM_1NN", "LPS", "TSBF", "TSF", "DTW_F", "EE", "BOSS", "ST", "FlatCOTE"]
idx = [classifiers.index(key) for key in subsample if key in classifiers]
plot_critical_difference(results_arr[:, idx], subsample, test="Nemenyi")
[6]:
(<Figure size 600x280 with 1 Axes>, <Axes: >)
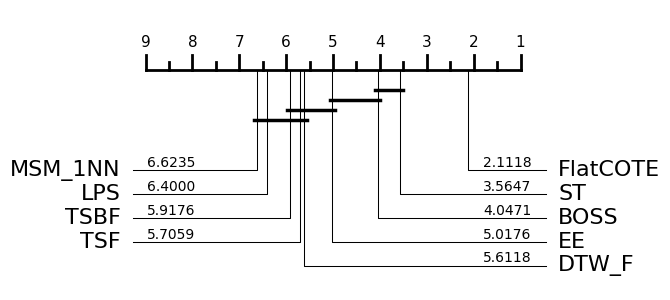
Note there are some small differences in averaged rank. This may be due to differences in how ties in rank were handled and missing resamples. The cliques are identical. Given that these results were generated in 2014/2015 using Java and Matlab was used to draw the diagrams, we think this is an acceptable reproduction. Subsequent to the 2015 bake off we switched to using pairwise Wilcoxon sign rank tests with the Holm correction. This creates slightly different cliques.
The great multivariate time series classification bake off, 2021¶
The multivariate bake off [2] launched a new archive and compared 11 classifiers on 26 multivariate TSC problems [5] over 30 resamples. The publication is available here
[7]:
from aeon.benchmarking.published_results import (
load_classification_bake_off_2021_results,
)
results_arr, datasets, classifiers = load_classification_bake_off_2021_results(
num_resamples=30, as_array=True
)
results_arr.shape
[7]:
(26, 11)
The datasets used in the 2021 bake off are as follows:
[8]:
datasets
[8]:
['ArticularyWordRecognition',
'AtrialFibrillation',
'BasicMotions',
'Cricket',
'DuckDuckGeese',
'EigenWorms',
'Epilepsy',
'EthanolConcentration',
'ERing',
'FaceDetection',
'FingerMovements',
'HandMovementDirection',
'Handwriting',
'Heartbeat',
'Libras',
'LSST',
'MotorImagery',
'NATOPS',
'PenDigits',
'PEMS-SF',
'PhonemeSpectra',
'RacketSports',
'SelfRegulationSCP1',
'SelfRegulationSCP2',
'StandWalkJump',
'UWaveGestureLibrary']
The classifiers used as follow:
[9]:
classifiers
[9]:
['CBOSS',
'CIF',
'DTW_D',
'DTW_I',
'gRSF',
'HIVE-COTEv1',
'ResNet',
'RISE',
'ROCKET',
'STC',
'TSF']
The results figures below shows the performance figures for accuracy, balanced accuracy, AUROC and F1.
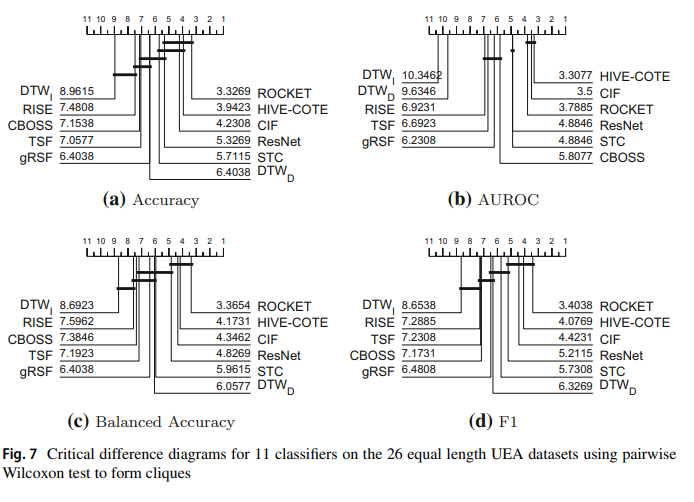
We can recreate the accuracy graph by plotting the loaded results like so:
[10]:
plot_critical_difference(results_arr, classifiers)
D:\CMP_Machine_Learning\Repositories\aeon\.venv\lib\site-packages\scipy\stats\_axis_nan_policy.py:600: UserWarning: Exact p-value calculation does not work if there are zeros. Switching to normal approximation.
return result_to_tuple(hypotest_fun_out(*samples, **kwds))
[10]:
(<Figure size 600x300 with 1 Axes>, <Axes: >)
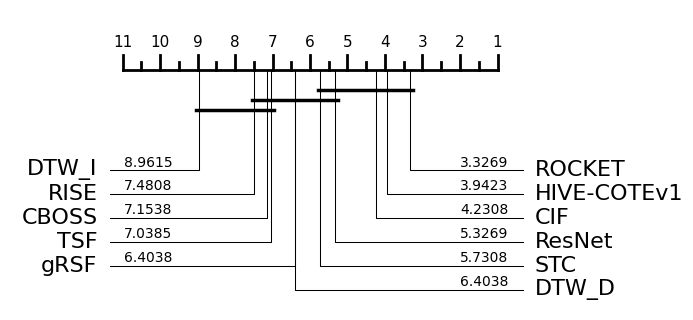
Note there are some differences in cliques due to slightly different methodology and tools used as mentioned previously.
Univariate bake off redux, 2023¶
In 2023 an update to the original 2017 bake off was produced, including the state of the art for the time [3]. This is the first bake off produced using the tools and estimators available in aeon. Published in 2024, the article is available here
34 algorithms were evaluated in total. 142 datasets were used for the top performing classifiers, and 112 for others. All results were averaged over 30 resamples.
Currently some estimators are missing. Only 112 datasets are available for all estimators, but in time we will upload the remaining datasets where applicable.
[11]:
from aeon.benchmarking.published_results import (
load_classification_bake_off_2023_results,
)
results_arr, datasets, classifiers = load_classification_bake_off_2023_results(
num_resamples=30, as_array=True
)
results_arr.shape
[11]:
(112, 34)
[12]:
datasets
[12]:
['ACSF1',
'Adiac',
'ArrowHead',
'Beef',
'BeetleFly',
'BirdChicken',
'BME',
'Car',
'CBF',
'Chinatown',
'ChlorineConcentration',
'CinCECGTorso',
'Coffee',
'Computers',
'CricketX',
'CricketY',
'CricketZ',
'Crop',
'DiatomSizeReduction',
'DistalPhalanxOutlineCorrect',
'DistalPhalanxOutlineAgeGroup',
'DistalPhalanxTW',
'Earthquakes',
'ECG200',
'ECG5000',
'ECGFiveDays',
'ElectricDevices',
'EOGHorizontalSignal',
'EOGVerticalSignal',
'EthanolLevel',
'FaceAll',
'FaceFour',
'FacesUCR',
'FiftyWords',
'Fish',
'FordA',
'FordB',
'FreezerRegularTrain',
'FreezerSmallTrain',
'GunPoint',
'GunPointAgeSpan',
'GunPointMaleVersusFemale',
'GunPointOldVersusYoung',
'Ham',
'HandOutlines',
'Haptics',
'Herring',
'HouseTwenty',
'InlineSkate',
'InsectEPGRegularTrain',
'InsectEPGSmallTrain',
'InsectWingbeatSound',
'ItalyPowerDemand',
'LargeKitchenAppliances',
'Lightning2',
'Lightning7',
'Mallat',
'Meat',
'MedicalImages',
'MiddlePhalanxOutlineCorrect',
'MiddlePhalanxOutlineAgeGroup',
'MiddlePhalanxTW',
'MixedShapesRegularTrain',
'MixedShapesSmallTrain',
'MoteStrain',
'NonInvasiveFetalECGThorax1',
'NonInvasiveFetalECGThorax2',
'OliveOil',
'OSULeaf',
'PhalangesOutlinesCorrect',
'Phoneme',
'PigAirwayPressure',
'PigArtPressure',
'PigCVP',
'Plane',
'PowerCons',
'ProximalPhalanxOutlineCorrect',
'ProximalPhalanxOutlineAgeGroup',
'ProximalPhalanxTW',
'RefrigerationDevices',
'Rock',
'ScreenType',
'SemgHandGenderCh2',
'SemgHandMovementCh2',
'SemgHandSubjectCh2',
'ShapeletSim',
'ShapesAll',
'SmallKitchenAppliances',
'SmoothSubspace',
'SonyAIBORobotSurface1',
'SonyAIBORobotSurface2',
'StarLightCurves',
'Strawberry',
'SwedishLeaf',
'Symbols',
'SyntheticControl',
'ToeSegmentation1',
'ToeSegmentation2',
'Trace',
'TwoLeadECG',
'TwoPatterns',
'UMD',
'UWaveGestureLibraryAll',
'UWaveGestureLibraryX',
'UWaveGestureLibraryY',
'UWaveGestureLibraryZ',
'Wafer',
'Wine',
'WordSynonyms',
'Worms',
'WormsTwoClass',
'Yoga']
[13]:
classifiers
[13]:
['Arsenal',
'BOSS',
'CIF',
'CNN',
'Catch22',
'DrCIF',
'EE',
'FreshPRINCE',
'HC1',
'HC2',
'Hydra-MR',
'Hydra',
'InceptionT',
'Mini-R',
'MrSQM',
'Multi-R',
'PF',
'RDST',
'RISE',
'ROCKET',
'RSF',
'RSTSF',
'ResNet',
'STC',
'ShapeDTW',
'Signatures',
'TDE',
'TS-CHIEF',
'TSF',
'TSFresh',
'WEASEL-D',
'WEASEL',
'cBOSS',
'1NN-DTW']
[14]:
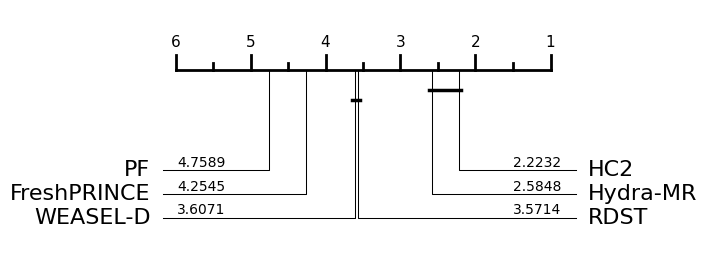
subsample = ["PF", "FreshPRINCE", "WEASEL-D", "RDST", "Hydra-MR", "HC2"]
idx = [classifiers.index(key) for key in subsample if key in classifiers]
plot_critical_difference(results_arr[:, idx], subsample)
[14]:
(<Figure size 600x250 with 1 Axes>, <Axes: >)
References¶
[1] Bagnall, A., Lines, J., Bostrom, A., Large, J. and Keogh, E., 2017. The great time series classification bake off: a review and experimental evaluation of recent algorithmic advances. Data mining and knowledge discovery, 31, pp.606-660.
[2] Ruiz, A.P., Flynn, M., Large, J., Middlehurst, M. and Bagnall, A., 2021. The great multivariate time series classification bake off: a review and experimental evaluation of recent algorithmic advances. Data Mining and Knowledge Discovery, 35(2), pp.401-449.
[3] Middlehurst, M., Schäfer, P. and Bagnall, A., 2024. Bake off redux: a review and experimental evaluation of recent time series classification algorithms. Data Mining and Knowledge Discovery, pp.1-74.
[4] Dau, H.A., Bagnall, A., Kamgar, K., Yeh, C.C.M., Zhu, Y., Gharghabi, S., Ratanamahatana, C.A. and Keogh, E., 2019. The UCR time series archive. IEEE/CAA Journal of Automatica Sinica, 6(6), pp.1293-1305.
[5] Bagnall, A., Dau, H.A., Lines, J., Flynn, M., Large, J., Bostrom, A., Southam, P. and Keogh, E., 2018. The UEA multivariate time series classification archive, 2018. arXiv preprint arXiv:1811.00075.
[6] Garcia, S. and Herrera, F., 2008. An Extension on” Statistical Comparisons of Classifiers over Multiple Data Sets” for all Pairwise Comparisons. Journal of machine learning research, 9(12).
Generated using nbsphinx. The Jupyter notebook can be found here.